Controller Adjustments¶
General Principle of Operation¶
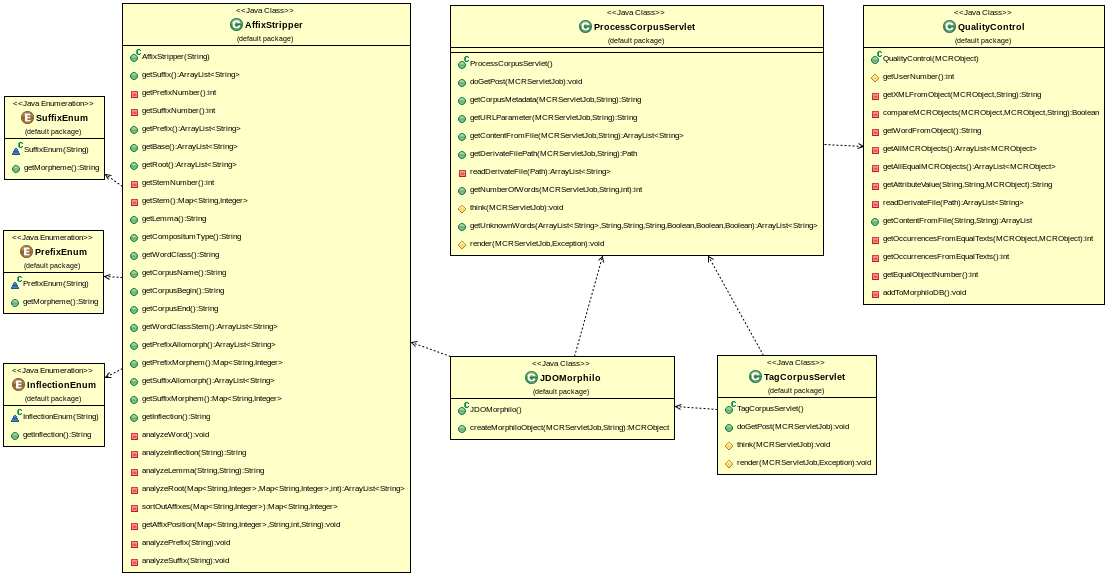
Figure 3: Morphilo UML Diagramm
Figure Figure 3: Morphilo UML Diagramm illustrates the dependencies of the five java classes that were integrated to add the morphilo functionality defined in the default package custom.mycore.addons.morphilo. The general principle of operation is the following. The handling of data search, upload, saving, and user authentification is fully left to the MyCoRe functionality that is completely implemented. The class ProcessCorpusServlet.java receives a request from the webinterface to process an uploaded file, i.e. a simple text corpus, and it checks if any of the words are available in the master database. All words that are not listed in the master database are written to an extra file. These are the words that have to be manually annotated. At the end, the servlet sends a response back to the user interface. In case of all words are contained in the master, an xml file is generated from the master database that includes all annotated words of the original corpus. Usually this will not be the case for larger textfiles. So if some words are not in the master, the user will get the response to initiate the manual annotation process.
The manual annotation process is processed by the class TagCorpusServlet.java, which will build a JDOM object for the first word in the extra file. This is done by creating an object of the JDOMorphilo.java class. This class, in turn, will use the methods of AffixStripper.java that make simple, but reasonable, suggestions on the word structure. This JDOM object is then given as a response back to the user. It is presented as a form, in which the user can make changes. This is necessary because the word structure algorithm of AffixStripper.java errs in some cases. Once the user agrees on the suggestions or on his or her corrections, the JDOM object is saved as an xml that is only searchable, visible, and changeable by the authenicated user (and the administrator), another file containing all processed words is created or updated respectively and the TagCorpusServlet.java servlet will restart until the last word in the extra list is processed. This enables the user to stop and resume her or his annotation work at a later point in time. The TagCorpusServlet will call methods from ProcessCorpusServlet.java to adjust the content of the extra files harboring the untagged words. If this file is empty, and only then, it is replaced by the file comprising all words from the original text file, both the ones from the master database and the ones that are annotated by the user, in an annotated xml representation.
Each time ProcessCorpusServlet.java is instantiated, it also instantiates QualityControl.java. This class checks if a new word can be transferred to the master database. The algorithm can be freely adopted to higher or lower quality standards. In its present configuration, a method tests at a limit of 20 different registered users agreeing on the annotation of the same word. More specifically, if 20 JDOM objects are identical except in the attribute field occurrences in the metadata node, the JDOM object becomes part of the master. The latter is easily done by changing the attribute creator from the user name to administrator in the service node. This makes the dataset part of the master database. Moreover, the occurrences attribute is updated by adding up all occurrences of the word that stem from different text corpora of the same time range.
Conceptualization¶
The controller component is largely specified and ready to use in some hundred or so java classes handling the logic of the search such as indexing, but also dealing with directories and files as saving, creating, deleting, and updating files. Moreover, a rudimentary user management comprising different roles and rights is offered. The basic technology behind the controller’s logic is the servlet. As such all new code has to be registered as a servlet in the web-fragment.xml (here the Apache Tomcat container).
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 | <servlet>
<servlet-name>ProcessCorpusServlet</servlet-name>
<servlet-class>custom.mycore.addons.morphilo.ProcessCorpusServlet</servlet-class>
</servlet>
<servlet-mapping>
<servlet-name>ProcessCorpusServlet</servlet-name>
<url-pattern>/servlets/object/process</url-pattern>
</servlet-mapping>
<servlet>
<servlet-name>TagCorpusServlet</servlet-name>
<servlet-class>custom.mycore.addons.morphilo.TagCorpusServlet</servlet-class>
</servlet>
<servlet-mapping>
<servlet-name>TagCorpusServlet</servlet-name>
<url-pattern>/servlets/object/tag</url-pattern>
</servlet-mapping>
\end{lstlisting}
|
Now, the logic has to be extended by the specifications. Some classes have to be added that take care of analyzing words (AffixStripper.java, InflectionEnum.java, SuffixEnum.java, PrefixEnum.java), extracting the relevant words from the text and checking the uniqueness of the text (ProcessCorpusServlet.java), make reasonable suggestions on the annotation (TagCorpusServlet.java), build the object of each annotated word (JDOMorphilo.java), and check on the quality by applying statistical models (QualityControl.java).
Implementation¶
Having taken a bird’s eye perspective in the previous chapter, it is now time to take a look at the specific implementation at the level of methods. Starting with the main servlet, ProcessCorpusServlet.java, the class defines four getter method:
- public String getURLParameter(MCRServletJob, String)
- public String getCorpusMetadata(MCRServletJob, String)
- public ArrayList<String> getContentFromFile(MCRServletJob, String)
- public Path getDerivateFilePath(MCRServletJob, String)
- public int getNumberOfWords(MCRServletJob job, String)
Since each servlet in MyCoRe extends the class MCRServlet, it has access to MCRServletJob, from which the http requests and responses can be used. This is the first argument in the above methods. The second argument of method (in 1.) specifies the name of an url parameter, i.e. the object id or the id of the derivate. The method returns the value of the given parameter. Typically MyCoRe uses the url to exchange these ids. The second method provides us with the value of a data field in the xml document. So the string defines the name of an attribute. getContentFromFile(MCRServletJob, String) returns the words as a list from a file when given the filename as a string. The getter listed in 4., returns the Path from the MyCoRe repository when the name of the file is specified. And finally, method (in 5.) returns the number of words by simply returning getContentFromFile(job, fileName).size().
There are two methods in every MyCoRe-Servlet that have to be overwritten, protected void render(MCRServletJob, Exception), which redirects the requests as POST or GET responds, and protected void think(MCRServletJob), in which the logic is implemented. Since the latter is important to understand the core idea of the Morphilo algorithm, it is displayed in full length in source code The overwritten think method.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 | protected void think(MCRServletJob job) throws Exception
{
this.job = job;
String dateFromCorp = getCorpusMetadata(job, "def.datefrom");
String dateUntilCorp = getCorpusMetadata(job, "def.dateuntil");
String corpID = getURLParameter(job, "objID");
String derivID = getURLParameter(job, "id");
//if NoW is 0, fill with anzWords
MCRObject helpObj = MCRMetadataManager.retrieveMCRObject(MCRObjectID.getInstance(corpID));
Document jdomDocHelp = helpObj.createXML();
XPathFactory xpfacty = XPathFactory.instance();
XPathExpression<Element> xpExp = xpfacty.compile("//NoW", Filters.element());
Element elem = xpExp.evaluateFirst(jdomDocHelp);
//fixes transferred morphilo data from previous stand alone project
int corpussize = getNumberOfWords(job, "");
if (Integer.parseInt(elem.getText()) != corpussize)
{
elem.setText(Integer.toString(corpussize));
helpObj = new MCRObject(jdomDocHelp);
MCRMetadataManager.update(helpObj);
}
//Check if the uploaded corpus was processed before
SolrClient slr = MCRSolrClientFactory.getSolrClient();
SolrQuery qry = new SolrQuery();
qry.setFields("korpusname", "datefrom", "dateuntil", "NoW", "id");
qry.setQuery("datefrom:" + dateFromCorp + " AND dateuntil:" + dateUntilCorp + " AND NoW:" + corpussize);
SolrDocumentList rslt = slr.query(qry).getResults();
Boolean incrOcc = true;
// if resultset contains only one, then it must be the newly created corpus
if (slr.query(qry).getResults().getNumFound() > 1)
{
incrOcc = false;
}
//match all words in corpus with morphilo (creator=administrator) and save all words that are not in morphilo DB in leftovers
ArrayList<String> leftovers = new ArrayList<String>();
ArrayList<String> processed = new ArrayList<String>();
leftovers = getUnknownWords(getContentFromFile(job, ""), dateFromCorp, dateUntilCorp, "", incrOcc, incrOcc, false);
//write all words of leftover in file as derivative to respective corpmeta dataset
MCRPath root = MCRPath.getPath(derivID, "/");
Path fn = getDerivateFilePath(job, "").getFileName();
Path p = root.resolve("untagged-" + fn);
Files.write(p, leftovers);
//create a file for all words that were processed
Path procWds = root.resolve("processed-" + fn);
Files.write(procWds, processed);
}
|
Using the above mentioned getter methods, the think-method assigns values to the object ID, needed to get the xml document that contains the corpus metadata, the file ID, and the beginning and starting dates from the corpus to be analyzed. Lines 10 through 22 show how to access a mycore object as an xml document, a procedure that will be used in different variants throughout this implementation. By means of the object ID, the respective corpus is identified and a JDOM document is constructed, which can then be accessed by XPath. The XPath factory instances are collections of the xml nodes. In the present case, it is save to assume that only one element of NoW is available (see corpus datamodel listing Corpus Data Model with maxOccurs=‘1’). So we do not have to loop through the collection, but use the first node named NoW. The if-test checks if the number of words of the uploaded file is the same as the number written in the document. When the document is initially created by the MyCoRe logic it was configured to be zero. If unequal, the setText(String) method is used to write the number of words of the corpus to the document.
Lines 25–36 reveal the second important ingredient, i.e. controlling the search engine. First, a solr client and a query are initialized. Then, the output of the result set is defined by giving the fields of interest of the document. In the case at hand, it is the id, the name of the corpus, the number of words, and the beginnig and ending dates. With setQuery it is possible to assign values to some or all of these fields. Finally, getResults() carries out the search and writes all hits to a SolrDocumentList (line 29). The test that follows is really only to set a Boolean encoding if the number of occurrences of that word in the master should be updated. To avoid multiple counts, incrementing the word frequency is only done if it is a new corpus.
In line 42 getUnknownWords(ArrayList, String, String, String, Boolean, Boolean, Boolean) is called and returned as a list of words. This method is key and will be discussed in depth below. Finally, lines 45–48 show how to handle file objects in MyCoRe. Using the file ID, the root path and the name of the first file in that path are identified. Then, a second file starting with untagged is created and all words returned from the getUnknownWords is written to that file. By the same token an empty file is created (in the last two lines of the think-method), in which all words that are manually annotated will be saved.
In a refactoring phase, the method getUnknownWords(ArrayList, String, String, String, Boolean, Boolean, Boolean) could be subdivided into three methods: for each Boolean parameter one. In fact, this method handles more than one task. This is mainly due to multiple code avoidance. In essence, an outer loop runs through all words of the corpus and an inner loop runs through all hits in the solr result set. Because the result set is supposed to be small, approximately between 10-20 items, efficiency problems are unlikely to cause a problem, although there are some more loops running through collection of about the same sizes. Since each word is identified on the basis of its projected word type, the word form, and the time range it falls into, it is these variables that have to be checked for existence in the documents. If not in the xml documents, null is returned and needs to be corrected. Moreover, user authentification must be considered. There are three different XPaths that are relevant.
- //service/servflags/servflag[@type=’createdby’] to test for the correct user
- //morphiloContainer/morphilo to create the annotated document
- //morphiloContainer/morphilo/w to set occurrences or add a link
As an illustration of the core functioning of this method, listing Mode of Operation of getUnknownWords Method is given.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 | public ArrayList<String> getUnknownWords(
ArrayList<String> corpus,
String timeCorpusBegin,
String timeCorpusEnd,
String wdtpe,
Boolean setOcc,
Boolean setXlink,
Boolean writeAllData) throws Exception
{
String currentUser = MCRSessionMgr.getCurrentSession().getUserInformation().getUserID();
ArrayList lo = new ArrayList();
for (int i = 0; i < corpus.size(); i++)
{
SolrClient solrClient = MCRSolrClientFactory.getSolrClient();
SolrQuery query = new SolrQuery();
query.setFields("w","occurrence","begin","end", "id", "wordtype");
query.setQuery(corpus.get(i));
query.setRows(50); //more than 50 items are extremely unlikely
SolrDocumentList results = solrClient.query(query).getResults();
Boolean available = false;
for (int entryNum = 0; entryNum < results.size(); entryNum++)
{
...
// update in MCRMetaDataManager
String mcrIDString = results.get(entryNum).getFieldValue("id").toString();
//MCRObjekt auslesen und JDOM-Document erzeugen:
MCRObject mcrObj = MCRMetadataManager.retrieveMCRObject(MCRObjectID.getInstance(mcrIDString));
Document jdomDoc = mcrObj.createXML();
...
//check and correction for word type
...
//checkand correction time: timeCorrect
...
//check if user correct: isAuthorized
...
XPathExpression<Element> xp = xpfac.compile("//morphiloContainer/morphilo/w", Filters.element());
//Iterates w-elements and increments occurrence attribute if setOcc is true
for (Element e : xp.evaluate(jdomDoc))
{
//wenn Rechte da sind und Worttyp nirgends gegeben oder gleich ist
if (isAuthorized && timeCorrect
&& ((e.getAttributeValue("wordtype") == null && wdtpe.equals(""))
|| e.getAttributeValue("wordtype").equals(wordtype))) // nur zur Vereinheitlichung
{
int oc = -1;
available = true;
try
{
//adjust occurrence Attribut
if (setOcc)
{
oc = Integer.parseInt(e.getAttributeValue("occurrence"));
e.setAttribute("occurrence", Integer.toString(oc + 1));
}
//write morphilo-ObjectID in xml of corpmeta
if (setXlink)
{
Namespace xlinkNamespace = Namespace.getNamespace("xlink", "http://www.w3.org/1999/xlink");
MCRObject corpObj = MCRMetadataManager.retrieveMCRObject(MCRObjectID.getInstance(getURLParameter(job, "objID")));
Document corpDoc = corpObj.createXML();
XPathExpression<Element> xpathEx = xpfac.compile("//corpuslink", Filters.element());
Element elm = xpathEx.evaluateFirst(corpDoc);
elm.setAttribute("href" , mcrIDString, xlinkNamespace);
}
mcrObj = new MCRObject(jdomDoc);
MCRMetadataManager.update(mcrObj);
QualityControl qc = new QualityControl(mcrObj);
}
catch(NumberFormatException except)
{
// ignore
}
}
}
if (!available) // if not available in datasets under the given conditions
{
lo.add(corpus.get(i));
}
}
return lo;
}
|
As can be seen from the functionality of listing Mode of Operation of getUnknownWords Method, getting the unknown words of a corpus, is rather a side effect for the equally named method. More precisely, a Boolean (line 47) is set when the document is manipulated otherwise because it is clear that the word must exist then. If the Boolean remains false (line 77), the word is put on the list of words that have to be annotated manually. As already explained above, the first loop runs through all words (corpus) and the following lines a solr result set is created. This set is also looped through and it is checked if the time range, the word type and the user are authorized. In the remainder, the occurrence attribute of the morphilo document can be incremented (setOcc is true) or/and the word is linked to the corpus meta data (setXlink is true). While all code lines are equivalent with what was explained in listing The overwritten think method, it suffices to focus on an additional name space, i.e. xlink has to be defined (line 60). Once the linking of word and corpus is set, the entire MyCoRe object has to be updated. This is done by the functionality of the framework (lines 67–69). At the end, an instance of QualityControl is created.
The class QualityControl is instantiated with a constructor depicted in listing Constructor of QualityControl.java.
private MCRObject mycoreObject;
/* Constructor calls method to carry out quality control, i.e. if at least 20
* different users agree 100% on the segments of the word under investigation
*/
public QualityControl(MCRObject mycoreObject) throws Exception
{
this.mycoreObject = mycoreObject;
if (getEqualObjectNumber() > 20)
{
addToMorphiloDB();
}
}
The constructor takes an MyCoRe object, a potential word candidate for the master data base, which is assigned to a private class variable because the object is used though not changed by some other java methods. More importantly, there are two more methods: getEqualNumber() and addToMorphiloDB(). While the former initiates a process of counting and comparing objects, the latter is concerned with calculating the correct number of occurrences from different, but not the same texts, and generating a MyCoRe object with the same content but with two different flags in the //service/servflags/servflag-node, i.e. createdby=’administrator’ and state=’published’. And of course, the occurrence attribute is set to the newly calculated value. The logic corresponds exactly to what was explained in listing The overwritten think method and will not be repeated here. The only difference are the paths compiled by the XPathFactory. They are
- //service/servflags/servflag[@type=’createdby’] and
- //service/servstates/servstate[@classid=’state’].
It is more instructive to document how the number of occurrences is calculated. There are two steps involved. First, a list with all mycore objects that are equal to the object which the class is instantiated with (mycoreObject in listing Constructor of QualityControl.java) is created. This list is looped and all occurrence attributes are summed up. Second, all occurrences from equal texts are substracted. Equal texts are identified on the basis of its meta data and its derivate.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 | /* returns number of Occurrences if Objects are equal, zero otherwise
*/
private int getOccurrencesFromEqualTexts(MCRObject mcrobj1, MCRObject mcrobj2) throws SAXException, IOException
{
int occurrences = 1;
//extract corpmeta ObjectIDs from morphilo-Objects
String crpID1 = getAttributeValue("//corpuslink", "href", mcrobj1);
String crpID2 = getAttributeValue("//corpuslink", "href", mcrobj2);
//get these two corpmeta Objects
MCRObject corpo1 = MCRMetadataManager.retrieveMCRObject(MCRObjectID.getInstance(crpID1));
MCRObject corpo2 = MCRMetadataManager.retrieveMCRObject(MCRObjectID.getInstance(crpID2));
//are the texts equal? get list of 'processed-words' derivate
String corp1DerivID = getAttributeValue("//structure/derobjects/derobject", "href", corpo1);
String corp2DerivID = getAttributeValue("//structure/derobjects/derobject", "href", corpo2);
ArrayList result = new ArrayList(getContentFromFile(corp1DerivID, ""));
result.remove(getContentFromFile(corp2DerivID, ""));
if (result.size() == 0) // the texts are equal
{
// extract occurrences of one the objects
occurrences = Integer.parseInt(getAttributeValue("//morphiloContainer/morphilo/w", "occurrence", mcrobj1));
}
else
{
occurrences = 0; //project metadata happened to be the same, but texts are different
}
return occurrences;
}
|
In this implementation, the ids from the corpmeta data model are accessed via the xlink attribute in the morphilo documents. The method getAttributeValue(String, String, MCRObject) does exactly the same as demonstrated earlier (see from line 60 on in listing Mode of Operation of getUnknownWords Method). The underlying logic is that the texts are equal if exactly the same number of words were uploaded. So all words from one file are written to a list (line 16) and words from the other file are removed from the very same list (line 17). If this list is empty, then the exact same number of words must have been in both files and the occurrences are adjusted accordingly. Since this method is called from another private method that only contains a loop through all equal objects, one gets the occurrences from all equal texts. For reasons of confirmability, the looping method is also given:
1 2 3 4 5 6 7 8 9 10 11 | private int getOccurrencesFromEqualTexts() throws Exception
{
ArrayList<MCRObject> equalObjects = new ArrayList<MCRObject>();
equalObjects = getAllEqualMCRObjects();
int occurrences = 0;
for (MCRObject obj : equalObjects)
{
occurrences = occurrences + getOccurrencesFromEqualTexts(mycoreObject, obj);
}
return occurrences;
}
|
Now, the constructor in listing Constructor of QualityControl.java reveals another method that rolls out an equally complex concatenation of procedures. As implied above, getEqualObjectNumber() returns the number of equally annotated words. It does this by falling back to another method from which the size of the returned list is calculated (getAllEqualMCRObjects().size()). Hence, we should care about getAllEqualMCRObjects(). This method really has the same design as int getOccurrencesFromEqualTexts() in listing Occurrence Extraction from Equal Texts (2). The difference is that another method (Boolean compareMCRObjects(MCRObject, MCRObject, String)) is used within the loop and that all equal objects are put into the list of MyCoRe objects that are returned. If this list comprises more than 20 entries, [1] the respective document will be integrated in the master data base by the process described above. The comparator logic is shown in listing Comparison of MyCoRe objects.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 | private Boolean compareMCRObjects(MCRObject mcrobj1, MCRObject mcrobj2, String xpath) throws SAXException, IOException
{
Boolean isEqual = false;
Boolean beginTime = false;
Boolean endTime = false;
Boolean occDiff = false;
Boolean corpusDiff = false;
String source = getXMLFromObject(mcrobj1, xpath);
String target = getXMLFromObject(mcrobj2, xpath);
XMLUnit.setIgnoreAttributeOrder(true);
XMLUnit.setIgnoreComments(true);
XMLUnit.setIgnoreDiffBetweenTextAndCDATA(true);
XMLUnit.setIgnoreWhitespace(true);
XMLUnit.setNormalizeWhitespace(true);
//differences in occurrences, end, begin should be ignored
try
{
Diff xmlDiff = new Diff(source, target);
DetailedDiff dd = new DetailedDiff(xmlDiff);
//counters for differences
int i = 0;
int j = 0;
int k = 0;
int l = 0;
// list containing all differences
List differences = dd.getAllDifferences();
for (Object object : differences)
{
Difference difference = (Difference) object;
//@begin,@end,... node is not in the difference list if the count is 0
if (difference.getControlNodeDetail().getXpathLocation().endsWith("@begin")) i++;
if (difference.getControlNodeDetail().getXpathLocation().endsWith("@end")) j++;
if (difference.getControlNodeDetail().getXpathLocation().endsWith("@occurrence")) k++;
if (difference.getControlNodeDetail().getXpathLocation().endsWith("@corpus")) l++;
//@begin and @end have different values: they must be checked if they fall right in the allowed time range
if ( difference.getControlNodeDetail().getXpathLocation().equals(difference.getTestNodeDetail().getXpathLocation())
&& difference.getControlNodeDetail().getXpathLocation().endsWith("@begin")
&& (Integer.parseInt(difference.getControlNodeDetail().getValue()) < Integer.parseInt(difference.getTestNodeDetail().getValue())) )
{
beginTime = true;
}
if (difference.getControlNodeDetail().getXpathLocation().equals(difference.getTestNodeDetail().getXpathLocation())
&& difference.getControlNodeDetail().getXpathLocation().endsWith("@end")
&& (Integer.parseInt(difference.getControlNodeDetail().getValue()) > Integer.parseInt(difference.getTestNodeDetail().getValue())) )
{
endTime = true;
}
//attribute values of @occurrence and @corpus are ignored if they are different
if (difference.getControlNodeDetail().getXpathLocation().equals(difference.getTestNodeDetail().getXpathLocation())
&& difference.getControlNodeDetail().getXpathLocation().endsWith("@occurrence"))
{
occDiff = true;
}
if (difference.getControlNodeDetail().getXpathLocation().equals(difference.getTestNodeDetail().getXpathLocation())
&& difference.getControlNodeDetail().getXpathLocation().endsWith("@corpus"))
{
corpusDiff = true;
}
}
//if any of @begin, @end ... is identical set Boolean to true
if (i == 0) beginTime = true;
if (j == 0) endTime = true;
if (k == 0) occDiff = true;
if (l == 0) corpusDiff = true;
//if the size of differences is greater than the number of changes admitted in @begin, @end ... something else must be different
if (beginTime && endTime && occDiff && corpusDiff && (i + j + k + l) == dd.getAllDifferences().size()) isEqual = true;
}
catch (SAXException e)
{
e.printStackTrace();
}
catch (IOException e)
{
e.printStackTrace();
}
return isEqual;
}
|
In this method, XMLUnit is heavily used to make all necessary node comparisons. The matter becomes more complicated, however, if some attributes are not only ignored, but evaluated according to a given definition as it is the case for the time range. If the evaluator and builder classes are not to be overwritten entirely because needed for evaluating other nodes of the xml document, the above solution appears a bit awkward. So there is potential for improvement before the production version is to be programmed.
XMLUnit provides us with a list of the differences of the two documents (see line 29). There are four differences allowed, that is, the attributes occurrence, corpus, begin, and end. For each of them a Boolean variable is set. Because any of the attributes could also be equal to the master document and the difference list only contains the actual differences, one has to find a way to define both, equal and different, for the attributes. This could be done by ignoring these nodes. Yet, this would not include testing if the beginning and ending dates fall into the range of the master document. Therefore the attributes are counted as lines 34 through 37 reveal. If any two documents differ in some of the four attributes just specified, then the sum of the counters (line 69) should not be greater than the collected differences by XMLUnit. The rest of the if-tests assign truth values to the respective Booleans. It is probably worth mentioning that if all counters are zero (lines 64–67) the attributes and values are identical and hence the Boolean has to be set explicitly. Otherwise the test in line 69 would fail.
Once quality control (explained in detail further down) has been passed, it is the user’s turn to interact further. By clicking on the option Manual tagging, the TagCorpusServlet will be callled. This servlet instantiates ProcessCorpusServlet to get access to the getUnknownWords-method, which delivers the words still to be processed and which overwrites the content of the file starting with untagged. For the next word in leftovers a new MyCoRe object is created using the JDOM API and added to the file beginning with processed. In line 16 of listing Manual Tagging Procedure, the previously defined entry mask is called, with which the proposed word structure could be confirmed or changed. How the word structure is determined will be shown later in the text.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 | ...
if (!leftovers.isEmpty())
{
ArrayList<String> processed = new ArrayList<String>();
//processed.add(leftovers.get(0));
JDOMorphilo jdm = new JDOMorphilo();
MCRObject obj = jdm.createMorphiloObject(job, leftovers.get(0));
//write word to be annotated in process list and save it
Path filePathProc = pcs.getDerivateFilePath(job, "processed").getFileName();
Path proc = root.resolve(filePathProc);
processed = pcs.getContentFromFile(job, "processed");
processed.add(leftovers.get(0));
Files.write(proc, processed);
//call entry mask for next word
tagUrl = prop.getBaseURL() + "content/publish/morphilo.xed?id=" + obj.getId();
}
else
{
//initiate process to give a complete tagged file of the original corpus
//if untagged-file is empty, match original file with morphilo
//creator=administrator OR creator=username and write matches in a new file
ArrayList<String> complete = new ArrayList<String>();
ProcessCorpusServlet pcs2 = new ProcessCorpusServlet();
complete = pcs2.getUnknownWords(
pcs2.getContentFromFile(job, ""), //main corpus file
pcs2.getCorpusMetadata(job, "def.datefrom"),
pcs2.getCorpusMetadata(job, "def.dateuntil"),
"", //wordtype
false,
false,
true);
Files.delete(p);
MCRXMLFunctions mdm = new MCRXMLFunctions();
String mainFile = mdm.getMainDocName(derivID);
Path newRoot = root.resolve("tagged-" + mainFile);
Files.write(newRoot, complete);
//return to Menu page
tagUrl = prop.getBaseURL() + "receive/" + corpID;
}
|
At the point where no more items are in leftsovers the getUnknownWords-method is called whereas the last Boolean parameter is set true. This indicates that the array list containing all available and relevant data to the respective user is returned as seen in the code snippet in listing ref{src:writeAll}.
...
// all data is written to lo in TEI
if (writeAllData && isAuthorized && timeCorrect)
{
XPathExpression<Element> xpath = xpfac.compile("//morphiloContainer/morphilo", Filters.element());
for (Element e : xpath.evaluate(jdomDoc))
{
XMLOutputter outputter = new XMLOutputter();
outputter.setFormat(Format.getPrettyFormat());
lo.add(outputter.outputString(e.getContent()));
}
}
...
The complete list (lo) is written to yet a third file starting with tagged and finally returned to the main project webpage.
The interesting question is now where does the word structure come from, which is filled in the entry mask as asserted above. In listing Manual Tagging Procedure line 7, one can see that a JDOM object is created and the method createMorphiloObject(MCRServletJob, String) is called. The string parameter is the word that needs to be analyzed. Most of the method is a mere application of the JDOM API given the data model in Conceptualization and listing Word Data Model. That means namespaces, elements and their attributes are defined in the correct order and hierarchy.
To fill the elements and attributes with text, i.e. prefixes, suffixes, stems, etc., a Hashmap – containing the morpheme as key and its position as value – are created that are filled with the results from an AffixStripper instantiation. Depending on how many prefixes or suffixes respectively are put in the hashmap, the same number of xml elements are created. As a final step, a valid MyCoRe id is generated using the existing MyCoRe functionality, the object is created and returned to the TagCorpusServlet.
Last, the analyses of the word structure will be considered. It is implemented in the AffixStripper.java file. All lexical affix morphemes and their allomorphs as well as the inflections were extracted from the Oxford English Dictionary and saved as enumerated lists (see the example in listing Enumeration Example for the Prefix over). The allomorphic items of these lists are mapped successively to the beginning in the case of prefixes (see listing Method to recognize prefixes, line 7) or to the end of words in the case of suffixes (see listing Cut-off mechanism for suffixes). Since each morphemic variant maps to its morpheme right away, it makes sense to use the morpheme and so implicitly keep the relation to its allomorph.
package custom.mycore.addons.morphilo;
public enum PrefixEnum {
...
over("over"), ufer("over"), ufor("over"), uferr("over"), uvver("over"), obaer("over"), ober("over)"), ofaer("over"),
ofere("over"), ofir("over"), ofor("over"), ofer("over"), ouer("over"),oferr("over"), offerr("over"), offr("over"), aure("over"),
war("over"), euer("over"), oferre("over"), oouer("over"), oger("over"), ouere("over"), ouir("over"), ouire("over"),
ouur("over"), ouver("over"), ouyr("over"), ovar("over"), overe("over"), ovre("over"),ovur("over"), owuere("over"), owver("over"),
houyr("over"), ouyre("over"), ovir("over"), ovyr("over"), hover("over"), auver("over"), awver("over"), ovver("over"),
hauver("over"), ova("over"), ove("over"), obuh("over"), ovah("over"), ovuh("over"), ofowr("over"), ouuer("over"), oure("over"),
owere("over"), owr("over"), owre("over"), owur("over"), owyr("over"), our("over"), ower("over"), oher("over"),
ooer("over"), oor("over"), owwer("over"), ovr("over"), owir("over"), oar("over"), aur("over"), oer("over"), ufara("over"),
ufera("over"), ufere("over"), uferra("over"), ufora("over"), ufore("over"), ufra("over"), ufre("over"), ufyrra("over"),
yfera("over"), yfere("over"), yferra("over"), uuera("over"), ufe("over"), uferre("over"), uuer("over"), uuere("over"),
vfere("over"), vuer("over"), vuere("over"), vver("over"), uvvor("over") ...
private String morpheme;
//constructor
PrefixEnum(String morpheme)
{
this.morpheme = morpheme;
}
//getter Method
public String getMorpheme()
{
return this.morpheme;
}
}
As can be seen in line 12 in listing Method to recognize prefixes, the morpheme is saved to a hash map together with its position, i.e. the size of the map plus one at the time being. In line 14 the analyzePrefix method is recursively called until no more matches can be made.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 | private Map<String, Integer> prefixMorpheme = new HashMap<String,Integer>();
...
private void analyzePrefix(String restword)
{
if (!restword.isEmpty()) //Abbruchbedingung fuer Rekursion
{
for (PrefixEnum prefEnum : PrefixEnum.values())
{
String s = prefEnum.toString();
if (restword.startsWith(s))
{
prefixMorpheme.put(s, prefixMorpheme.size() + 1);
//cut off the prefix that is added to the list
analyzePrefix(restword.substring(s.length()));
}
else
{
analyzePrefix("");
}
}
}
}
|
The recognition of suffixes differs only in the cut-off direction since suffixes occur at the end of a word. Hence, line 14 in listing Method to recognize prefixes reads in the case of suffixes.
analyzeSuffix(restword.substring(0, restword.length() - s.length()));
It is important to note that inflections are suffixes (in the given model case of Middle English morphology) that usually occur at the very end of a word, i.e. after all lexical suffixes, only once. It follows that inflections have to be recognized at first without any repetition. So the procedure for inflections can be simplified to a substantial degree as listing Method to recognize inflections shows.
private String analyzeInflection(String wrd)
{
String infl = "";
for (InflectionEnum inflEnum : InflectionEnum.values())
{
if (wrd.endsWith(inflEnum.toString()))
{
infl = inflEnum.toString();
}
}
return infl;
}
Unfortunately the embeddedness problem prevents a very simple algorithm. Embeddedness occurs when a lexical item is a substring of another lexical item. To illustrate, the suffix ion is also contained in the suffix ation, as is ent in ment, and so on. The embeddedness problem cannot be solved completely on the basis of linear modelling, but for a large part of embedded items one can work around it using implicitly Zipf’s law, i.e. the correlation between frequency and length of lexical items. The longer a word becomes, the less frequent it will occur. The simplest logic out of it is to assume that longer suffixes (measured in letters) are preferred over shorter suffixes because it is more likely tha the longer the suffix string becomes, the more likely it is one (as opposed to several) suffix unit(s). This is done in listing Method to workaround embeddedness, whereas the inner class sortedByLengthMap returns a list sorted by length and the loop from line 17 onwards deletes the respective substrings.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 | private Map<String, Integer> sortOutAffixes(Map<String, Integer> affix)
{
Map<String,Integer> sortedByLengthMap = new TreeMap<String, Integer>(new Comparator<String>()
{
@Override
public int compare(String s1, String s2)
{
int cmp = Integer.compare(s1.length(), s2.length());
return cmp != 0 ? cmp : s1.compareTo(s2);
}
}
);
sortedByLengthMap.putAll(affix);
ArrayList<String> al1 = new ArrayList<String>(sortedByLengthMap.keySet());
ArrayList<String> al2 = al1;
Collections.reverse(al2);
for (String s2 : al1)
{
for (String s1 : al2)
if (s1.contains(s2) && s1.length() > s2.length())
{
affix.remove(s2);
}
}
return affix;
}
|
Finally, the position of the affix has to be calculated because the hashmap in line 12 in listing Method to recognize prefixes does not keep the original order for changes taken place in addressing the affix embeddedness (listing Method to workaround embeddedness). Listing Method to determine position of the affix depicts the preferred solution. The recursive construction of the method is similar to private void analyzePrefix(String) (listing Method to recognize prefixes) only that the two affix types are handled in one method. For that, an additional parameter taking the form either suffix or prefix is included.
private void getAffixPosition(Map<String, Integer> affix, String restword, int pos, String affixtype)
{
if (!restword.isEmpty()) //Abbruchbedingung fuer Rekursion
{
for (String s : affix.keySet())
{
if (restword.startsWith(s) && affixtype.equals("prefix"))
{
pos++;
prefixMorpheme.put(s, pos);
//prefixAllomorph.add(pos-1, restword.substring(s.length()));
getAffixPosition(affix, restword.substring(s.length()), pos, affixtype);
}
else if (restword.endsWith(s) && affixtype.equals("suffix"))
{
pos++;
suffixMorpheme.put(s, pos);
//suffixAllomorph.add(pos-1, restword.substring(s.length()));
getAffixPosition(affix, restword.substring(0, restword.length() - s.length()), pos, affixtype);
}
else
{
getAffixPosition(affix, "", pos, affixtype);
}
}
}
}
To give the complete word structure, the root of a word should also be provided. In listing Method to determine roots a simple solution is offered, however, considering compounds as words consisting of more than one root.
private ArrayList<String> analyzeRoot(Map<String, Integer> pref, Map<String, Integer> suf, int stemNumber)
{
ArrayList<String> root = new ArrayList<String>();
int j = 1; //one root always exists
// if word is a compound several roots exist
while (j <= stemNumber)
{
j++;
String rest = lemma;
for (int i=0;i<pref.size();i++)
{
for (String s : pref.keySet())
{
//if (i == pref.get(s))
if (rest.length() > s.length() && s.equals(rest.substring(0, s.length())))
{
rest = rest.substring(s.length(),rest.length());
}
}
}
for (int i=0;i<suf.size();i++)
{
for (String s : suf.keySet())
{
//if (i == suf.get(s))
if (s.length() < rest.length() && (s.equals(rest.substring(rest.length() - s.length(), rest.length()))))
{
rest = rest.substring(0, rest.length() - s.length());
}
}
}
root.add(rest);
}
return root;
}
The logic behind this method is that the root is the remainder of a word when all prefixes and suffixes are substracted. So the loops run through the number of prefixes and suffixes at each position and substract the affix. Really, there is some code doubling with the previously described methods, which could be eliminated by making it more modular in a possible refactoring phase. Again, this is not the concern of a prototype. Line 9 defines the initial state of a root, which is the case for monomorphemic words. The lemma is defined as the wordtoken without the inflection. Thus listing Method to determine lemma reveals how the class variable is calculated
/*
* Simplification: lemma = wordtoken - inflection
*/
private String analyzeLemma(String wrd, String infl)
{
return wrd.substring(0, wrd.length() - infl.length());
}
The constructor of AffixStripper calls the method analyzeWord() whose only job is to calculate each structure element in the correct order (listing Method to determine lemma. All structure elements are also provided by getters.
private void analyzeWord()
{
//analyze inflection first because it always occurs at the end of a word
inflection = analyzeInflection(wordtoken);
lemma = analyzeLemma(wordtoken, inflection);
analyzePrefix(lemma);
analyzeSuffix(lemma);
getAffixPosition(sortOutAffixes(prefixMorpheme), lemma, 0, "prefix");
getAffixPosition(sortOutAffixes(suffixMorpheme), lemma, 0, "suffix");
prefixNumber = prefixMorpheme.size();
suffixNumber = suffixMorpheme.size();
wordroot = analyzeRoot(prefixMorpheme, suffixMorpheme, getStemNumber());
}
To conclude, the Morphilo implementation as presented here, aims at fulfilling the task of a working prototype. It is important to note that it neither claims to be a very efficient nor a ready software program to be used in production. However, it marks a crucial milestone on the way to a production system. At some listings sources of improvement were made explicit; at others no suggestions were made. In the latter case this does not imply that there is no potential for improvement. Once acceptability tests are carried out, it will be the task of a follow up project to identify these potentials and implement them accordingly.
Notes
| [1] | This number is somewhat arbitrary. It is inspired by the sample size n in t-distributed data. |